
Viruses: potential genetic vectors for transfer between species
“Jumping genes” are elements of the genome which are capable of being transmitted from one species to another. To understand how they behave, researchers from the Évolution, génomes, comportement et écologie (Evolution, genomes, behaviour and ecology) laboratory (EGCE - Université Paris-Saclay, CNRS, IRD) study viruses and examine them as potential genetic vectors.
Transposable elements, also known as “jumping genes”, are DNA sequences which can move from one chromosome to another. They are present in all organisms and represent about 50% of the human genome. They are, nevertheless, considered to be genomic parasites, as once they become part of the DNA, they cannot be dislodged from it. An exacerbating factor is that their presence is harmful to the host as they are a source of genetic disease. The organism cannot get rid of them. Only genetic mutations combined with natural selection manage to inactivate and break them down.
The transferable elements embedded in the host's DNA are naturally transferred from generation to generation via reproduction. This is referred to as vertical transmission. However, they can also be transferred between different species. For example, identical DNA sequences are found in the rhodnius haematophagous bug and the squirrel monkey. This as yet unexplained phenomenon is called horizontal transmission. Clément Gilbert, a researcher at the ECGE laboratory (Université Paris-Saclay, CNRS, IRD) is conducting several studies to understand this process.
Inequality in the frequency of transmission between transposable elements
Generally, transposable elements are classified into two groups according to their mode of horizontal transmission: group 1 includes those that use “copy and paste” mode and group 2 includes those that “cut and paste”. In the case of the first group, an enzyme transcribes the transposable element into RNA (copy) and then reverse transcribes it into DNA (paste). In the second group, an enzyme cuts the DNA strand (cut), extracts the transposable element from it and reintegrates it elsewhere in the genome (paste).
Curiously, these two modes of transfer are not observed with the same frequency in organisms, as the mode used by group 1 is more widely used than that of group 2. To shed some light on this, Clément Gilbert and his team have measured the constraints acting on transposable elements during different types of transfers in nearly 307 species of vertebrates.
The transposable elements are generally all subject to natural selection and undergo mutations which inactivate them. For those in group 2, their change of position within the host genome takes place independently of their activity since the enzyme responsible for transporting them does not recognise this state. Over time, mutations then repeat and the rate of inactive copies of the element increases within the genome. Conversely, the enzyme has to recognise elements in group 1 in order to be able to “copy and paste” them. They are transferred because they are active. Over time, an accumulation of active elements can be observed which then leads on to a wider transmission of the elements in group 1. This is called the purifying selection.
![Circular phylogenetic tree showing the 975 horizontal transfers of transposable elements (blue lines [group 1] and red lines [group 2]) between the 307 vertebrates studied.](/sites/default/files/styles/large/public/2020-12/Article%20Transfert%20horizontaux%20Fig1_0.jpg?itok=pa3KA1r6)
Inequality in the frequency of horizontal transmission between species
It is even more surprising that horizontal transmission does not take place at the same rate between all species. To pinpoint the cause of this phenomenon, Clément Gilbert’s team have measured the number of horizontal transfers in the 307 vertebrate species. The researchers analysed the genome of each species to identify transposable elements, then selected just their horizontal transfers.
They counted no less than 975 occurrences of horizontal transfers, the vast majority of which were in teleost fish. This surprising result made the team wonder why the fish are targeted so much. “Several hypotheses have been put forward, but we do not have a definitive answer yet. This may be that there is an excess of transfer in teleost fish or a lack of transfer in other vertebrate species. The external fertilization of fish increasing the chances of foreign DNA integration is also a possibility,” suggests Clément Gilbert.
Viruses as a source of genetic transfer between species
But how exactly does a foreign gene integrate into an organism? At the moment, this type of horizontal transfer remains a mystery. However, Clément Gilbert believes that perhaps parasites and viruses play a key role. “Viruses replicate within the cells of their host. As a result, their genetic material is very close to that of their host. This could lead to exchanges of DNA between the two,” explains the researcher.
Some viruses can be genetic vectors between two host species. However several conditions must be met. The virus must be transmissible between these two species, it must not kill them and then there must be a vertical transfer so that the transposable element remains within the second species. “After integrating the transposable element of the first host, and if it does not kill its host and is not released into the environment, the virus will be transmitted to a second host. If it reaches the germ cells, it will be able to penetrate, replicate and transpose the element of the first host,” explains Clément Gilbert.
To support this hypothesis, the researcher has set up the TransVir project with the aim of studying the potential of certain viruses to be vectors for horizontal transfers. The project has been developed together with the laboratoire Écologie et biologie des interactions (Ecology and Interaction Biology Laboratory - Université de Poitiers, CNRS) and the Institut de recherche sur la biologie de l’insecte (Institute for Research on Insect Biology – Université de Tours, CNRS) and has received € 240 000-worth of funding from the Agence nationale de recherche (ANR - National Research Agency) to last around five years (December 2015 – March 2021).
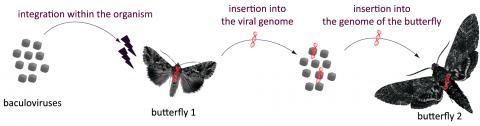
Together, the researchers were interested in horizontal transfers in insects involving baculoviruses. The results obtained have quickly shown that on average 5% of baculovirus genomes carry a transposable element from butterfly caterpillars. This confirms the first step in the transfer process from a host to a virus.
Over the next few years, Clément Gilbert would like to renew this collaboration in order to focus on a new ANR project and study the second stage of the process - namely the transfer from the virus to a second host. He also plans to study the potential of extracellular vesicles as vectors for horizontal transfers. These organelles, which ensure communication between cells, could also be a source of gene transfer between species.
*The baculoviruses infect butterfly 1. When the virus replicates, the transposable elements of butterfly 1 are inserted into the viral genome. This transfers them to butterfly 2. During the infection of butterfly 2, the transposable elements of butterfly 1 carried by the virus transpose into the genome of butterfly 2, resulting in a horizontal transfer.
- Gilbert, C., Peccoud, J., & Cordaux, R. Transposable Elements and the Evolution of Insects. Annual Review of Entomology, 66 (2020).
- Zhang, H. H., Peccoud, J., Zhang, X. G., & Gilbert, C. Horizontal transfer and evolution of transposable elements in vertebrates. Nature communications, 11(1), 1-10 (2020).